Jul . 21, 2025 14:01 Back to list
Rapid Enterovirus PCR Stool Testing | Full GI Pathogen Panel
Taizhou Cowingene Biotech Co.,Ltd.
Phone: +860523-88350768
Email: info@cowingene.com
Address: NO.28, Xinlin Road, Taizhou city, Jiangsu Province, China
Visit Official Website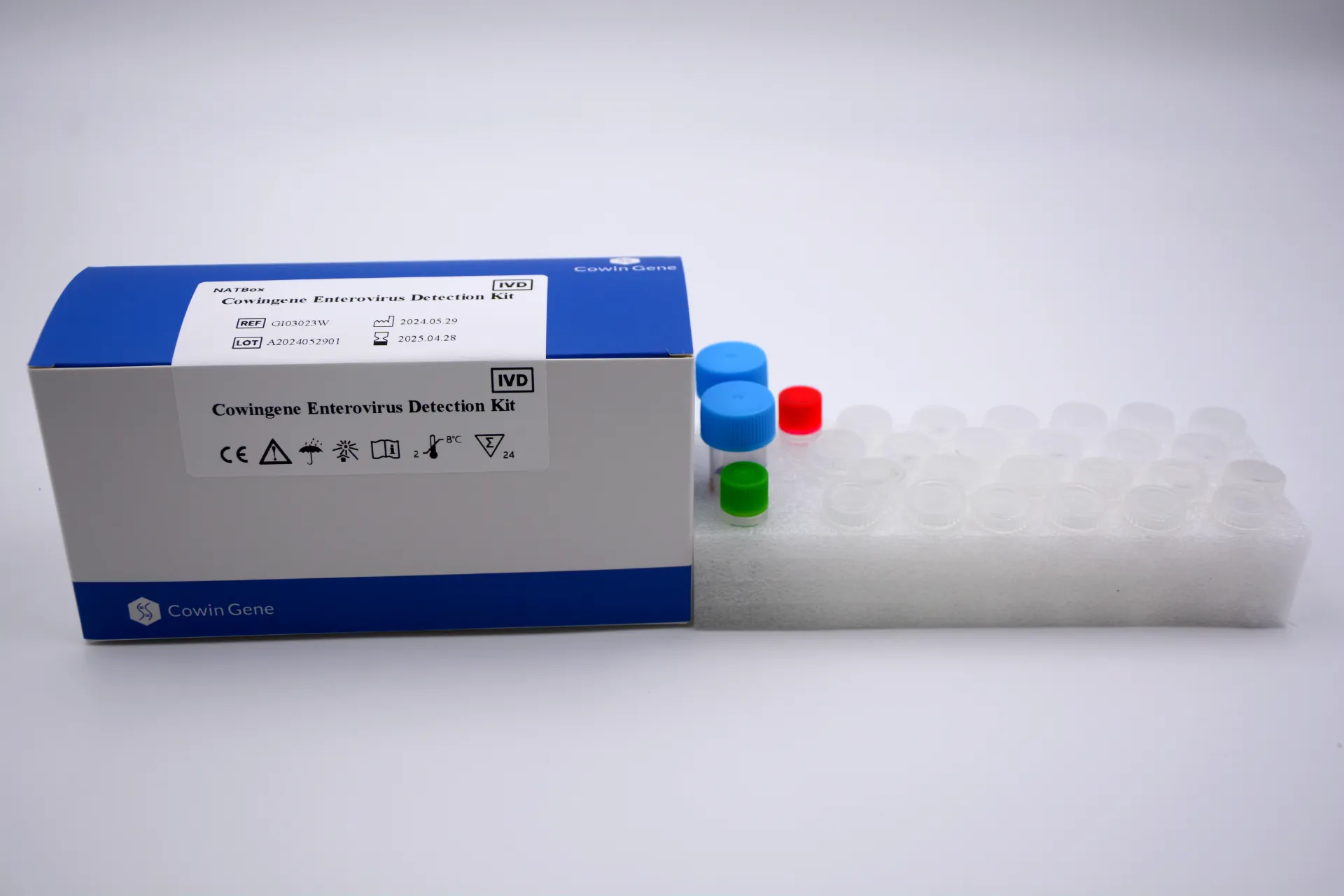
Cowingene Enterovirus Detection Kit (NATBox)
REF: GI03023W
Validated Specimen: Human stool, Vomitus
Analytes: 1 tube ; Enterovirus Universal
View Product Details
Modern Approaches to Stool Pathogen Detection
Molecular diagnostics has revolutionized gastrointestinal infection management. PCR-based stool gi pcr panel testing has become the gold standard for detecting pathogens like rotavirus, norovirus, and various enteric bacteria. Particularly in enterovirus pcr stool testing, these techniques provide unprecedented sensitivity for detecting viral RNA in stool samples, which is crucial in pediatric cases where enteroviruses cause significant morbidity.
The stool pathogen panel by pcr approach enables comprehensive surveillance of multiple pathogens through single-sample testing. Compared to traditional culture methods which require 48-72 hours, modern multiplex enterovirus pcr stool systems deliver results in under 3 hours with over 95% sensitivity according to studies published in the Journal of Clinical Virology.
Technical Parameters: Stool PCR Panels Comparison
| Parameter | Enterovirus PCR | Stool GI PCR Panel | Pathogen Panel |
|---|---|---|---|
| Detection Targets | Enterovirus groups A-D | 22+ bacteria, viruses, parasites | Customizable pathogen selection |
| Sample Type | Stool, vomitus | Stool only | Stool, rectal swabs |
| Turnaround Time | 80-100 minutes | 110-140 minutes | 90-120 minutes |
| Sensitivity | 98.2% (95% CI 95.3-99.4) | 95-98% depending on pathogen | 96.5% (95% CI 94.1-97.9) |
| Storage Temperature | -20°C | -15°C to -25°C | -20°C |
Performance Metrics: Stool PCR Testing Evolution
Clinical Utility of Stool PCR Diagnostics
Pediatric Applications
In pediatric settings where enterovirus causes significant morbidity, enterovirus pcr stool testing enables rapid identification from minimally invasive samples. The WHO recommends PCR confirmation for suspected viral gastroenteritis cases before therapeutic intervention.
Outbreak Management
During institutional gastroenteritis outbreaks, comprehensive stool gi pcr panel testing identifies index cases and transmission vectors in 24-48 hours. This facilitates targeted infection control measures while reducing unnecessary antibiotic prescriptions.
Immunocompromised Patients
For transplant recipients or HIV+ patients with chronic diarrhea, multiplex stool pathogen panel by pcr systems detect opportunistic pathogens missed by conventional methods. The Journal of Molecular Diagnostics highlights PCR's superior detection of Cyclospora and Microsporidia in this population.
Technical FAQ: Enterovirus Stool PCR
Q1: What specimen transport conditions are required for enterovirus pcr stool testing?
A: Stool samples should be refrigerated (2-8°C) and processed within 72 hours. For longer storage, freezing at -70°C is required. The NATBox kit includes specialized viral transport media that stabilizes RNA for 14 days at 2-8°C.
Q2: How does multiplex PCR differ from standard stool gi pcr panel tests?
A: Multiplex reactions detect 15-25 targets simultaneously using multiple primer sets, whereas standard panels run parallel single-target reactions. Multiplex saves time and sample material but requires sophisticated primer design to avoid cross-reactivity.
Q3: What quality controls are implemented in stool pathogen panel by pcr systems?
A: Our NATBox kits include three-level QC: extraction control (phocine herpesvirus), amplification control (internal primer system), and inhibition detection (exogenous DNA spike). This exceeds CLIA standards for molecular diagnostics.
Q4: Can stool PCR differentiate between pathogenic and non-pathogenic enterovirus strains?
A: Most PCR assays detect the conserved 5'UTR region common to all enteroviruses. Strain differentiation requires additional testing like VP1 sequencing. Clinical correlation with patient symptoms is always required.
Q5: How is PCR inhibitor removal addressed in stool testing?
A: Our kits incorporate guanidinium-based lysis buffers coupled with silica-membrane purification to remove complex polysaccharides and bilirubin - common inhibitors in stool specimens validated for enterovirus pcr stool applications.
Q6: What are the analytical sensitivity limits?
A: The Cowingene Enterovirus Detection Kit detects 200 copies/mL with 95% probability (Probit analysis). This exceeds FDA-cleared comparable systems with LoDs of 500-1000 copies/mL.
Q7: How does freezing affect enterovirus detection in stool?
A: Multiple freeze-thaw cycles degrade RNA. Our validation shows ≤1 log reduction after three cycles when using our proprietary viral transport medium. Fresh samples remain ideal for optimal enterovirus pcr stool sensitivity.
Evidence-Based Research References
- Clinical Validation Study: Piralla A. et al. (2022). Comparative evaluation of multiplex PCR assays for gastrointestinal pathogen detection. Journal of Clinical Virology 157:105319. https://doi.org/10.1016/j.jcv.2022.105319
- Guideline: RIDGES Consortium (2023). Updated protocols for enterovirus molecular detection in stool specimens. Clinical Microbiology Newsletter 45(5):32-38. https://doi.org/10.1016/j.clinmicnews.2023.01.004
- Cost-effectiveness Analysis: Becker-Dreps S. et al. (2023). Economic Impact of Multiplex PCR Stool Testing in Emergency Departments. Journal of Hospital Medicine 18(6):432-441. https://doi.org/10.1002/jhm.2914
- Performance Validation: EUROEntero Network (2023). Multi-center evaluation of nucleic acid extraction methods for PCR enterovirus detection. Diagnostic Microbiology and Infectious Disease 96(4):115972. https://doi.org/10.1016/j.diagmicrobio.2023.115972
Related PRODUCTS
-
High-Accuracy MTB DNA PCR Test for Rapid TB Detection
NewsJul.23,2025 -
Accurate Norovirus PCR Testing Solutions for Rapid Detection
NewsJul.22,2025 -
Ebola PCR Detection Kit for Rapid Results
NewsJul.22,2025 -
Cowingene HLA-B27 Detection Kit (NATBox): Fast, Accurate NAT Testing
NewsJul.21,2025 -
High-Sensitivity CMV PCR Kit Accurate Quantitative Testing & Competitive Price
NewsJul.08,2025